Difference between revisions of "Data Analysis - Starting Out"
| Line 107: | Line 107: | ||
==Naming Conventions for PARAM files== | ==Naming Conventions for PARAM files== | ||
| + | The desired kinematics have already been specified for the EMC and X>1 experiments. Some minor changes may be applied, but for the most part they are not changing. Different angle settings and momentum settings are used to target different physics. Detector calibrations, such as the calorimeter and DC calibrations, change with these kinematic changes. For this reason, '''we name our calibration PARAM files based on the kinematic setting they apply to'''.<br>For example, all the SRC runs in 2018 were taken at 8 degrees and 9.8GeV central momentum. We thus name our calibration files as we normally would, but before the sp18 suffix, we add the '''8deg_m9p8_sp18'''. For the SHMS calorimeter calibration this would then be: | ||
| + | <code>PARAM/SHMS/CAL/pcal_calib_8deg_m9p8_sp18.param</code> | ||
| + | * The SHMS calorimeter p (for SHMS) followed by the angle and momentum setting and the run period suffix. Here the decimal has been replaced with a '''p''' and the '''m''' indicates minus momentum. This is to distinguish between positron CSB settings and regular data taking. | ||
| + | There were no conflicts with this system for 2018 and 2019 running. This being the case, the standard.database is read per-run-number. If there was a problem with this nomenclature, as long as the files had different names, standard.database could sort it out. | ||
| + | There is nothing special with this naming scheme other than the ease of finding calibration files for specific settings. This is especially useful when debugging an issue with a specific group of runs. | ||
==Running a SCRIPT== | ==Running a SCRIPT== | ||
Revision as of 21:55, 26 September 2021
Starting Out
The Overview section should be viewed before starting this section. (Currently under construction)
The purpose of this section is to give new members of the XEM group hands-on experience with 'hcana' and the 'replay'. A bare-bones git branch named practice has been added to the hallc_replay_XEM repository.
If none of this makes sense, make sure you have followed along with the getting started software section.
It is recommended all practice be performed in a separate directory on your /group/ disk such as:
/group/c-xem2/<CUE_username>/PRACTICE/
The practice branch is used for practice, and is not meant to be your long-term analysis branch. You should be on the master branch to get the most up-to-date calibrations. If performing a new calibration or contributing to the hallc_replay_XEM, new work should be added to a new_feature_branch and merged into your origin new_feature_branch. This is followed by a push to mrcmor100 master. Refer to the git help and software overview sections to familiarize yourself with this workflow.
Objectives
- Learn to setup your practice directory
- Set up all hallc_replay_XEM directories
- Know what standard.kinematics and standard.database files do
- Learn to include parameters in hcana
- Naming conventions of parameters in hcana
- Naming conventions of PARAM files in hallc_replay_XEM
- Run your first replay and explore the output
- Print the value of a parameter from hcana to the console
You should not make pull requests to Casey for your practice branch. If you want him to check that you're doing something correct, push the local changes to your origin and notify Casey via Slack.
Setting up the Replay
First you must have forked the hallc_replay_XEM repository from mrcmor100. This is outlined in the getting started - software section.
- In /group/c-xem2/<CUE_username>/PRACTICE/ disk location:
git clone git@github.com:<username>/hallc_replay_XEM.git
- Username should be your github username, CUE_username is your JLab username.
- Follow along with the software section to setup git with ssh on JLab computers.
- cd into the hallc_replay_XEM directory
git submodule initgit submodule updategit fetch origin practicegit checkout practice
Now you should have all the submodules used in the hallc_replay_XEM directory and you should be on the practice branch. Check this by typing
git branchin the hallc_replay_XEM directory.
Add Directories to the Replay
The output of hcana goes to two locations: ROOTfiles and REPORT_OUTPUT directories. These are saved on the /volatile/ disk to prevent using the /group/ location inappropriately. These directories are symbolically linked to the /volatile/ directories. The linked /volatile/ directory names can have any name you like, but the directory names in hallc_replay_XEM must be ROOTfiles and REPORT_OUTPUT.
mkdir /volatile/hallc/xem2/<CUE_username>/<desired_practice_rootfiles>mkdir /volatile/hallc/xem2/<CUE_username>/<desired_practice_report_output>ln -s /volatile/hallc/xem2/<CUE_username>/<desired_practice_rootfiles> ROOTfilesln -s /volatile/hallc/xem2/<CUE_username>/<desired_practice_report_output> REPORT_OUTPUT
The output is further separated into more subdirectories to separate files for the calibration, production, and spectrometer type. These are as follows:
- ROOTfiles/SHMS/
- CALIBRATION
- PRODUCTION
- TIMING
- SCALERS
- ROOTfiles/HMS/
- CALIBRATION
- PRODUCTION
- TIMING
- SCALERS
The CALIBRATION directories are used for storing the output of calibration SCRIPTS. The TIMING directories are used to hold the output of the timing SCRIPTS in SCRIPTS/TIMING, etc. These calibrations, timing, production, and scaler scripts are explained in more detail on other pages of the data analysis pages.
We must also point the hallc_replay_XEM to the raw EVIO data files. This is done by adding the appropriate /raw directories:
ln -s /cache/mss/hallc/spring17/raw raw-sp18ln -s /cache/mss/hallc/jpsi-007/raw raw-sp19
These locations are the front face of the tape library. Files may not always be available here, and must be retrieved by the jcache utility. For more information refer to the scicomp documentation and the file structure and farm sections of getting started to use the jcace utility.
Standard dot what?
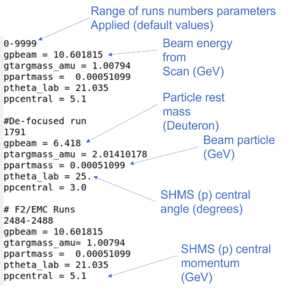
The standard.kinematics file tells the analyzer information about the beam energy, particle, spectrometer angle, target mass, and central momentum. The p and g are used to denote SHMS (p) and global(g) variables. If this were the HMS standard.kinematics file, the prefix p would be changed to h (HMS). There are two main kinematics files used for the XEM experiment, one for each spectrometer.
DBASE/HMS/standard.kinematicsDBASE/SHMS/standard.kinematics
The notation for run number range is common throughout all PARAM files for hcana.
- A single run is typed before the select parameters for that run.
- A dash - between two runs is the run range over which the parameters are applied.
- The file is loaded from top to bottom, so parameters at the beginning are overwritten by later parameters.
- This also applies to loading a file with parameters of the same name. PARAM files loaded later will overwrite previous parameters. Refer to the viewing hcana parameter section for an example.
- The # sign indicates a comment that is not read by hcana.
Note: "#include" is a special set of characters that hcana recognizes.
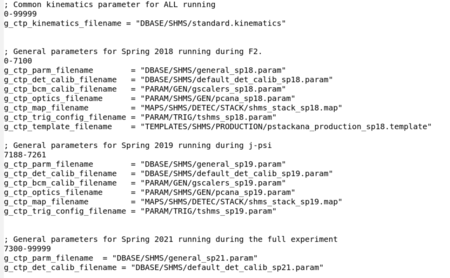
Aside from target and beam-specific parameters, hcana also needs to take in information about the detector geometry, data cuts, and detector calibrations. These parameters are loaded in from the standard.database file.
DBASE/HMS/standard.databaseDBASE/SHMS/standard.database
Several strings are defined at the head of standard.database. These strings given in quotes are the names of files to load separately into hcana. Note that the spaces have no meaning in the input to hcana parameters, and the run-ranges above could have been listed as '2484 - 2488'.
Notice in this file there are three separate run ranges. Since the X>1 data has been taken in two separate time periods (and a third to be held in 2022), there are different PARAM files to load for different configurations. Generally, in the X>1 and EMC analysis, parameters relevant for spring of 2018 have the sp18 suffix. Files associated with spring 2019 sp19 suffix. Some parameters did not change between 2018 and 2019, and are not expected to change between now and the 2022 running. Those files do not have a suffix.
Refer to the general parameters and spectrometer general parameters for information on parameters contained within these files.
Brief description of each part filename imported via the standard.database file.
- g_ctp_parm_filename - General parameters. typically detector geometry and nominal acceptance cuts.
- g_ctp_det_calib_filename - default calibrations for each detector.
- g_ctp_bcm_calib_filename - Default scaler calibrations for specific run periods. Most of these are overwritten in specific run ranges based on beam current calibrations. Refer to the BCM calibration section.
- g_ctp_optics_filename - A set of matrix elements for reconstruction of events at the target. These are general parameters, but more specific ones are typically loaded for the spring 2018 runs due to the optics issues encountered. Refer to the optics section of data analysis.
- g_ctp_map_filename - This is an important map of all signals in the readout controllers (ROCs). This can change from period-to-period, but typically doesn't change. This does not change within run periods.
- g_ctp_trig_config_filename - This file typically doesn't change from period-to-period, but with the trigger changes between 2018 and 2019 ours did. This was to remove some trigger legs and add some helicity information.
- g_ctp_template_filename - This is to load specific template files used to form the REPORT_OUTPUT/*.report files. This should remain the same within a run period so that report files do not change over the course of the run. More relevant branches have been added over time making these report files more useful.
HCANA Naming Conventions
hallc_replay_XEM is a framework of calibration files, replay scripts and database files that point the hallc analyzer (hcana) to the appropriate data files. Some of the recognized naming in hcana has been outlined in the previous section. Those are recapped here and more information is added. The name of parameters must match that of the source code. You can ensure this is done by 'grepping' the source code of hcana.
- Parameters typed after a run number and before another run number are applied to that specific run number.
- A dash - between two runs is the run range over which the parameters below are applied.
- The file is loaded from top to bottom, so parameters at the beginning are overwritten by later parameters.
- Generally, parameters begin with a string and are separated by a parenthesis.
- Spaces do not matter here.
- Variables are loaded in using the DBRequest functionailty in the code of hcana. In this way, the variable type is given. hcana will typically tell you if it is trying to load a decimal number into an integer.
- Arrays of variables can span multiple lines and are separated by commas. hcana typically knows how large of an array to expect, so it may give you an error or warning while running.
- This also applies to loading a file with parameters of the same name. PARAM files loaded later will overwrite previous parameters. Refer to the viewing hcana parameter section for an example.
- The # sign indicates a comment that is not read by hcana and can be used on the same line as a parameter specification.
- The ; also indicates a comma and can be used on the same line as a parameter specification.
- The "#include" is a special set of characters that hcana recognizes. hcana will automatically load the contents of that PARAM file if found.
Naming Conventions for PARAM files
The desired kinematics have already been specified for the EMC and X>1 experiments. Some minor changes may be applied, but for the most part they are not changing. Different angle settings and momentum settings are used to target different physics. Detector calibrations, such as the calorimeter and DC calibrations, change with these kinematic changes. For this reason, we name our calibration PARAM files based on the kinematic setting they apply to.
For example, all the SRC runs in 2018 were taken at 8 degrees and 9.8GeV central momentum. We thus name our calibration files as we normally would, but before the sp18 suffix, we add the 8deg_m9p8_sp18. For the SHMS calorimeter calibration this would then be:
PARAM/SHMS/CAL/pcal_calib_8deg_m9p8_sp18.param
- The SHMS calorimeter p (for SHMS) followed by the angle and momentum setting and the run period suffix. Here the decimal has been replaced with a p and the m indicates minus momentum. This is to distinguish between positron CSB settings and regular data taking.
There were no conflicts with this system for 2018 and 2019 running. This being the case, the standard.database is read per-run-number. If there was a problem with this nomenclature, as long as the files had different names, standard.database could sort it out. There is nothing special with this naming scheme other than the ease of finding calibration files for specific settings. This is especially useful when debugging an issue with a specific group of runs.