Difference between revisions of "TriDAS + JANA2 (+ERSAP)"
| (33 intermediate revisions by the same user not shown) | |||
| Line 9: | Line 9: | ||
== Meetings == | == Meetings == | ||
| + | * [[TriDAS/JANA2/ERSAP 2023-03-08 | 2023-03-08]] | ||
| + | * [[TriDAS/JANA2/ERSAP 2023-03-01 | 2023-03-01]] | ||
| + | * [[TriDAS/JANA2/ERSAP 2023-02-22 | 2023-02-22]] | ||
| + | * [[TriDAS/JANA2/ERSAP 2023-02-15 | 2023-02-15]] | ||
* [[TriDAS/JANA2/ERSAP 2023-02-08 | 2023-02-08]] | * [[TriDAS/JANA2/ERSAP 2023-02-08 | 2023-02-08]] | ||
* [[TriDAS/JANA2/ERSAP 2023-01-27 | 2023-01-27]] | * [[TriDAS/JANA2/ERSAP 2023-01-27 | 2023-01-27]] | ||
| Line 25: | Line 29: | ||
| step 2: || Run TriDAS w/ JANA2 for 3x3 prototype in Hall-B CH || 2/9 || 2/14 || INFN(Stefano, Carmello), Sergey B., EPSCI(David, Nathan, Cissie) | | step 2: || Run TriDAS w/ JANA2 for 3x3 prototype in Hall-B CH || 2/9 || 2/14 || INFN(Stefano, Carmello), Sergey B., EPSCI(David, Nathan, Cissie) | ||
|- | |- | ||
| − | | step 3: || | + | | step 3: || Run TriDAS w/ JANA2 for 5x5 prototype in Hall-B CH || 2/14 || 2/16 || INFN(Stefano, Carmello), Sergey B., EPSCI(David, Nathan, Cissie) |
|- | |- | ||
| − | | step 4: || | + | | step 4: || Install 5x5 calorimeter in Hall-D|| 2/17 || 2/27 || INFN |
|- | |- | ||
| − | | step 5: || | + | | step 5: || Run TriDAS w/ JANA2 for 5x5 prototype using cosmic and beam triggers|| 2/27 || 3/5 || INFN(Stefano, Carmello), Sergey B., EPSCI(David, Nathan, Cissie) |
|- | |- | ||
| − | | step 6: || | + | | step 6: || Integrate AI-based real-time processing via JANA2 plugin || 3/6 || 3/13 || Cristiano, Cissie |
|- | |- | ||
| − | | step 7: || Collect final data || 3/15 || 3/20 || | + | | step 7: || TriDAS -> ERSAP -> JANA2 stack processing || 3/13 || 3/15 || INFN(Stefano, Carmello), Sergey B., EPSCI(Vardan, Carl, David) |
| + | |- | ||
| + | | step 8: || Collect final data || 3/15 || 3/20 || | ||
|} | |} | ||
| Line 80: | Line 86: | ||
* ERSAP iterface to JANA2 | * ERSAP iterface to JANA2 | ||
* JANA2 Beam trigger and Cosmic Ray trigger | * JANA2 Beam trigger and Cosmic Ray trigger | ||
| + | |||
| + | == Instructions for Starting TriDAS DAQ == | ||
| + | There are two separate DAQ systems currently configured. One for the 5x5 calorimeter in the Hall-D beamline and the other for the 3x3 calorimeter in the Hall-B counting house. Currently, we run only one at a time. If both need to be run simultaneously, then contact Sergey Boiarinov. Instructions for running the DAQ for either system are below with differences for the calorimeters indicated. | ||
| + | |||
| + | IMPORTANT: When starting a new run '''''ALWAYS''''' take CODA all the back to the ''Configure'' stage. If it is not reconfigured for each run then the firmware is known to report incorrect channel numbers! | ||
| + | |||
| + | Start up CODA RunControl | ||
| + | # Log into the ''clasrun'' account on clondaq5: | ||
| + | > ssh clasrun@clondaq5 | ||
| + | |||
| + | # Launch VNC viewer for the session that should already be running | ||
| + | # (you will need to know the password) | ||
| + | > vncviewer localhost:5901 | ||
| + | |||
| + | # If the CODA RunControl GUI is not already up, launch it with the | ||
| + | # second command below. For an absolutely clean start, kill all | ||
| + | # existing processes first: | ||
| + | > killall runcontrol rcServer rocs | ||
| + | > runcontrol -rocs | ||
| + | |||
| + | # You need to get the DAQ up to the stage where it is ready | ||
| + | # to have the "Prestart" button pressed. If ''runcontrol'' is already | ||
| + | # running, then make sure to click 'Abort' first so you can then press | ||
| + | # 'Configure'. | ||
| + | # n.b. trigger files are located in /usr/clas12/release/1.4.0_streaming/params/trigger/STREAMING | ||
| + | - Click 'Connect' | ||
| + | - Click 'Configure' | ||
| + | for 5x5 calorimeter in Hall-D choose: 'HALLDFDC1_OLD_DUMMY_DAQ3' | ||
| + | for 3x3 calorimeter in Hall-B choose: 'TEST2_OLD_DUMMY' | ||
| + | - Click 'Download' | ||
| + | for 5x5 calorimeter beam in Hall-D choose: 'STREAMING/halldfdc1_streaming_tridas.trg ' | ||
| + | for 5x5 calorimeter cosmics in Hall-D choose: 'STREAMING/halldfdc1_streaming_tridas_cosmic.trg' | ||
| + | for 3x3 calorimeter cosmics in Hall-B choose: 'STREAMING/test2_streaming_tridas.trg' | ||
| + | |||
| + | Start up TriDAS processes | ||
| + | # Open a new terminal and log into the ''streamdaq'' account on clondaq3 for the 5x5 | ||
| + | # or clondaq5 for the 3x3 (yes, that sounds backwards!): | ||
| + | > ssh streamdaq@clondaq3 # for 5x5 calorimeter in Hall-D | ||
| + | or | ||
| + | > ssh streamdaq@clondaq5 # for 3x3 calorimeter in Hall-B | ||
| + | |||
| + | # Make sure the RunTSC process is running. This is often left running | ||
| + | # in a hidden tmux window. If it is already running then skip this part. | ||
| + | # If it is not running do the following: | ||
| + | > tmux ls | ||
| + | |||
| + | # (find the right window number. Probably the latest one. e.g. "6") | ||
| + | > tmux att -t 6 | ||
| + | > RunTSC -x /var/run/user/10350/tsc.sock | ||
| + | |||
| + | # Type ''ctl-b d'' to detach from the tmux window and leave it running | ||
| + | # in the background. | ||
| + | |||
| + | # Start the tridas shell | ||
| + | > tscsh --socket /var/run/user/10350/tsc.sock | ||
| + | |||
| + | # At the tridas shell prompt enter one of the following: | ||
| + | # --- for 5x5 calorimeter cosmics in Hall-D: | ||
| + | > init /home/streamdaq/tridas/run/datacard_repo/tridas_datacard2022-clondaq3-cosmic-M5EICv1.json | ||
| + | |||
| + | # --- for 5x5 calorimeter beam in Hall-D: | ||
| + | > init /home/streamdaq/tridas/run/datacard_repo/tridas_datacard2022-clondaq3-beam-M5EICv1.json | ||
| + | |||
| + | # --- for 3x3 calorimeter in Hall-B: | ||
| + | > init /home/streamdaq/tridas/run/datacard_repo/tridas_datacard2022-clondaq5-M5EICv1.json | ||
| + | |||
| + | # Then, for all run types, enter: | ||
| + | > configure | ||
| + | > start | ||
| + | |||
| + | Start Run | ||
| + | # Back in the VNC window on the RunControl GUI: | ||
| + | - Click 'Prestart', connection should be established from test2vtp-s2 to clondaq5 port 7002 | ||
| + | - Click 'Go' - data should be streaming to clondaq5 port 7002 | ||
| + | |||
| + | End Run | ||
| + | # To End the run: | ||
| + | # In ''tscsh'' tridas shell | ||
| + | > stop | ||
| + | > reset | ||
| + | |||
| + | # In VNC RunControl GUI | ||
| + | - Click 'Stop' | ||
| + | - Click 'Abort' | ||
| + | |||
| + | # n.b. remember to click 'Configure' and then 'Download' if you wish to take another run! | ||
| + | |||
| + | == Analyzing the Data == | ||
| + | Output data files are written to a directory/filename format specified in the ''datacard.json'' configuration in the "EM" section by the "PT_NAME_TEMPLATE" field. For the cosmic runs in the Hall-B Counting house, they will have names like: | ||
| + | /data/tridas/EIC_3x3_R00000235_F00000003.pt | ||
| + | |||
| + | You can check the file format is good using PTFileAnalyser like this: | ||
| + | cd /usr/clas12/streaming/work/2022.12.15.JANA2_TriDAS/tridas-core | ||
| + | ./build/tools/PTFileAnalyser --ptfile /data/tridas/EIC_3x3_R00000255_F00000000.pt | ||
| + | |||
| + | For the 3x3 calorimeter, you can use JANA and the ''EIC_3x3'' plugin to create a ROOT file with some histograms and trees containing the data. Note that this requires the ''PTFileSource'' and ''EIC_3x3_cosmics_trigger'' plugins also be attached. The former for reading ''.pt'' file and translation table and the latter for identifying tracks and triggers in the event. The following will create a file named ''EIC_3x3.root''. | ||
| + | |||
| + | jana -Pplugins=PTFileSource,EIC_3x3_cosmics_trigger,EIC_3x3 /data/tridas/EIC_3x3_R00000321_F00000000.pt | ||
| + | |||
| + | Note: the source for the ''EIC_3x3'' plugin can be found on the web [https://github.com/andrea-celentano/streamingReco/tree/master/src/plugins/EIC_3x3 here] and on clondaq5 in the directory: | ||
| + | /usr/clas12/streaming/work/2022.12.15.JANA2_TriDAS/streamingReco/src/plugins/EIC_3x3 | ||
| + | |||
| + | The other plugins are also in subdirectories of ''plugins''. | ||
== Software == | == Software == | ||
| − | |||
| − | + | [[File:20230223_streamingReco_Software.png|frameless|500px]] | |
| + | |||
| + | Work is being done using the ''streamdaq'' account on ''clondaq5''. | ||
| + | |||
| + | NOTE: The .bashrc file for the streamdaq account has been updated to point to the current software versions. The following is currently '''''not''''' needed to complete the environment setup. | ||
| − | Some software built specifically for this test can be found in the '''''/usr/clas12/streaming/work/2022.12.15.JANA2_TriDAS''''' directory. Not all of it is placed there though. | + | <s>source /usr/clas12/streaming/work/2022.12.15.JANA2_TriDAS/setenv.sh</s> |
| + | |||
| + | Some software built specifically for this test can be found in the '''''/usr/clas12/streaming/work/2022.12.15.JANA2_TriDAS''''' directory. Not all of it is placed there though. Commands used to build most of the packages are recorded in the /usr/clas12/streaming/work/2022.12.15.JANA2_TriDAS/README file. | ||
Some notes describing how these pieces fit together from 2020 when this was first set up can be seen here: | Some notes describing how these pieces fit together from 2020 when this was first set up can be seen here: | ||
| Line 93: | Line 207: | ||
=== Compiler === | === Compiler === | ||
| − | * 9.3.1 (from | + | * 9.3.1 (from devtoolset-9) |
* C++17 standard | * C++17 standard | ||
=== TriDAS === | === TriDAS === | ||
| − | This is the TriDAS source and is checked out on the latest ''master''. | + | This is the TriDAS source and is checked out on the latest ''master''. It was needed since ''TriggerInterface.h'' (and possibly other files) needed by the ''jtridas'' plugin were not present in the binary distribution that was installed. |
* /usr/clas12/streaming/work/2022.12.15.JANA2_TriDAS/tridas-core | * /usr/clas12/streaming/work/2022.12.15.JANA2_TriDAS/tridas-core | ||
| + | |||
| + | === tscsh === | ||
| + | The tridas shell ''tscsh'' has also been built from the latest master. It is built in the directory below which automatically added to ''PATH'' in ''.bashrc''. | ||
| + | * /usr/clas12/streaming/work/2022.12.15.JANA2_TriDAS/tscell/build | ||
=== jtridas === | === jtridas === | ||
This is the TriDAS plugin that interfaces to [https://jeffersonlab.github.io/JANA2/ JANA2]. TriDAS will load this plugin and, in turn, this will instantiate a JANA2 ''JApplication'' that will then load all of the JANA2 plugins. It is responsible for interfacing TriDAS and JANA2. | This is the TriDAS plugin that interfaces to [https://jeffersonlab.github.io/JANA2/ JANA2]. TriDAS will load this plugin and, in turn, this will instantiate a JANA2 ''JApplication'' that will then load all of the JANA2 plugins. It is responsible for interfacing TriDAS and JANA2. | ||
* /usr/clas12/streaming/work/2022.12.15.JANA2_TriDAS/jtridas | * /usr/clas12/streaming/work/2022.12.15.JANA2_TriDAS/jtridas | ||
| + | |||
| + | NOTE: This plugin needs to be at least symlinked in the directory where tridas looks for plugins as specified in the ''datacard.json'' file. (This is the "PLUGINS_DIR" field in the "TCPU" section.) The following does this for this beam test: | ||
| + | |||
| + | ln -s /usr/clas12/streaming/work/2022.12.15.JANA2_TriDAS/jtridas/build_vscode/libTrigJANA.so /usr/clas12/streaming/work/2022.12.15.JANA2_TriDAS/tridas-core/install/trigger_plugins | ||
=== streamingReco === | === streamingReco === | ||
| Line 111: | Line 233: | ||
=== ROOT === | === ROOT === | ||
ROOT was built specifically for this test using the above compiler and C++ standard | ROOT was built specifically for this test using the above compiler and C++ standard | ||
| − | * /usr/clas12/streaming/ | + | * /usr/clas12/streaming/root/root-6.26.10 |
=== CCDB === | === CCDB === | ||
| − | CCDB is | + | CCDB is the JLab ''Calibrations Constants Database''. Version 1.06.09 from github is checked out here. |
| − | * /usr/clas12/streaming/ | + | * /usr/clas12/streaming/ccdb_versions/v1.06.09 |
=== JANA2 === | === JANA2 === | ||
The latest tagged version of JANA2 v2.0.8 was built for this test. Note that the install directory is given below, but the full source can be found one directory up. | The latest tagged version of JANA2 v2.0.8 was built for this test. Note that the install directory is given below, but the full source can be found one directory up. | ||
* /usr/clas12/streaming/JANA2-v2.0.8/install | * /usr/clas12/streaming/JANA2-v2.0.8/install | ||
| + | |||
| + | === git === | ||
| + | The ''tridas-core'' CMakeLists.txt runs git internally with the ''-C'' option which is only available in newer versions of git. Thus, the latest version (2.39.1 as of now) was built here. | ||
| + | * /usr/clas12/streaming/git/git-2.39.1 | ||
| + | |||
| + | === cmake === | ||
| + | The ''tridas-core'' CMakeLists.txt requires version 3.20 or higher of cmake. A precompiled version was downloaded and installed here. | ||
| + | * /usr/clas12/streaming/cmake-3.26.0-rc1-linux-x86_64 | ||
| + | |||
| + | === libssh2 === | ||
| + | TriDAS requires Libssh2 version 1.9. A previous build of this from 2020 exists in ''/usr/clas12/streaming/libssh2-1.9.0'' but was apparently built as a static library which caused complaints when linking that we need to rebuild it with -fPIC. This was done and the shared library version placed here. (The older build with the static library was left alone). | ||
| + | * /usr/clas12/streaming/work/2022.12.15.JANA2_TriDAS/libssh2 | ||
=== VSCode === | === VSCode === | ||
| Line 129: | Line 263: | ||
== Useful links == | == Useful links == | ||
| + | * [https://docs.google.com/spreadsheets/d/1K16PELnJtM83MXIPXrQF_-7zDzThNGIPDXxP3MrOi4I/edit?usp=sharing Run Spreadsheet] | ||
* [https://baltig.infn.it/tridas/tridas-core/-/blob/jlab2021/packages/DataFormats/s_dataformat_clas12.hpp New data format ](better suited for VTP) | * [https://baltig.infn.it/tridas/tridas-core/-/blob/jlab2021/packages/DataFormats/s_dataformat_clas12.hpp New data format ](better suited for VTP) | ||
* [https://github.com/andrea-celentano/streamingReco streamingReco GitHub repository] | * [https://github.com/andrea-celentano/streamingReco streamingReco GitHub repository] | ||
* [https://github.com/andrea-celentano/jtridas jtridas GitHub Repository] | * [https://github.com/andrea-celentano/jtridas jtridas GitHub Repository] | ||
* Data located at: /data/tridas/EIC* | * Data located at: /data/tridas/EIC* | ||
Latest revision as of 15:04, 13 March 2023
This is information related to beam tests done in support of the TriDAS/JANA2/ERSAP SRO tests in Spring 2023 in Hall-D.
Overview
The goal of this test is to read out a small matrix of scintillator bars using VTP modules in Streaming ReadOut (SRO) mode. The bars have dimensions 2x2x20 cm and are side by side and closely packed (no space between them). The matrix will be positioned so the long dimension will be aligned with the beam direction. Cosmic rays will therefore be largely perpendicular to that.
The initial phase will focus on a 3x3 matrix while a 5x5 matrix will be used for the beam test. The beam test will take place in March, but development using cosmic rays will occur in the months leading up to that.
Meetings
Timetable of Activities
| Activity | start date | end date | responsible | |
|---|---|---|---|---|
| step 1: | Build existing software against current TriDAS and JANA2 | 2/6 | 2/8 | EPSCI (David) |
| step 2: | Run TriDAS w/ JANA2 for 3x3 prototype in Hall-B CH | 2/9 | 2/14 | INFN(Stefano, Carmello), Sergey B., EPSCI(David, Nathan, Cissie) |
| step 3: | Run TriDAS w/ JANA2 for 5x5 prototype in Hall-B CH | 2/14 | 2/16 | INFN(Stefano, Carmello), Sergey B., EPSCI(David, Nathan, Cissie) |
| step 4: | Install 5x5 calorimeter in Hall-D | 2/17 | 2/27 | INFN |
| step 5: | Run TriDAS w/ JANA2 for 5x5 prototype using cosmic and beam triggers | 2/27 | 3/5 | INFN(Stefano, Carmello), Sergey B., EPSCI(David, Nathan, Cissie) |
| step 6: | Integrate AI-based real-time processing via JANA2 plugin | 3/6 | 3/13 | Cristiano, Cissie |
| step 7: | TriDAS -> ERSAP -> JANA2 stack processing | 3/13 | 3/15 | INFN(Stefano, Carmello), Sergey B., EPSCI(Vardan, Carl, David) |
| step 8: | Collect final data | 3/15 | 3/20 |
| Capability Focus | start date | end date |
|---|---|---|
| JANA2 plugins - TriDAS | 2/6 | 2/10 |
| JANA2 plugins - CDAQ | 2/13 | 2/17 |
| CDAQ + JANA2 | 2/20 | 2/24 |
| TriDAS + JANA2 | 2/27 | 3/3 |
| ERSAP + TriDAS | 3/6 | 3/10 |
| ERSAP + CDAQ | 3/13 | 3/17 |
| Accelerator Down | 3/20 | |
Project Phases
Phase 1: mid-Feb.
- Cosmic rays (3x3)
- VTP readout with TriDAS
- JANA2 Cosmic Ray trigger
Phase 2: end-Feb.
- Cosmic rays (5x5)
- VTP readout with TriDAS
- JANA2 Cosmic Ray trigger
Phase 3: mid-Mar.
- Beam Test (5x5)
- VTP readout with TriDAS
- JANA2 Beam trigger and Cosmic Ray trigger
Phase 4: end-Mar.
- Beam Test (5x5)
- VTP readout with ERSAP (+ TriDAS?)
- ERSAP iterface to JANA2
- JANA2 Beam trigger and Cosmic Ray trigger
Instructions for Starting TriDAS DAQ
There are two separate DAQ systems currently configured. One for the 5x5 calorimeter in the Hall-D beamline and the other for the 3x3 calorimeter in the Hall-B counting house. Currently, we run only one at a time. If both need to be run simultaneously, then contact Sergey Boiarinov. Instructions for running the DAQ for either system are below with differences for the calorimeters indicated.
IMPORTANT: When starting a new run ALWAYS take CODA all the back to the Configure stage. If it is not reconfigured for each run then the firmware is known to report incorrect channel numbers!
Start up CODA RunControl
# Log into the clasrun account on clondaq5:
> ssh clasrun@clondaq5
# Launch VNC viewer for the session that should already be running
# (you will need to know the password)
> vncviewer localhost:5901
# If the CODA RunControl GUI is not already up, launch it with the
# second command below. For an absolutely clean start, kill all
# existing processes first:
> killall runcontrol rcServer rocs
> runcontrol -rocs
# You need to get the DAQ up to the stage where it is ready
# to have the "Prestart" button pressed. If runcontrol is already
# running, then make sure to click 'Abort' first so you can then press
# 'Configure'.
# n.b. trigger files are located in /usr/clas12/release/1.4.0_streaming/params/trigger/STREAMING
- Click 'Connect'
- Click 'Configure'
for 5x5 calorimeter in Hall-D choose: 'HALLDFDC1_OLD_DUMMY_DAQ3'
for 3x3 calorimeter in Hall-B choose: 'TEST2_OLD_DUMMY'
- Click 'Download'
for 5x5 calorimeter beam in Hall-D choose: 'STREAMING/halldfdc1_streaming_tridas.trg '
for 5x5 calorimeter cosmics in Hall-D choose: 'STREAMING/halldfdc1_streaming_tridas_cosmic.trg'
for 3x3 calorimeter cosmics in Hall-B choose: 'STREAMING/test2_streaming_tridas.trg'
Start up TriDAS processes
# Open a new terminal and log into the streamdaq account on clondaq3 for the 5x5
# or clondaq5 for the 3x3 (yes, that sounds backwards!):
> ssh streamdaq@clondaq3 # for 5x5 calorimeter in Hall-D
or
> ssh streamdaq@clondaq5 # for 3x3 calorimeter in Hall-B
# Make sure the RunTSC process is running. This is often left running
# in a hidden tmux window. If it is already running then skip this part.
# If it is not running do the following:
> tmux ls
# (find the right window number. Probably the latest one. e.g. "6")
> tmux att -t 6
> RunTSC -x /var/run/user/10350/tsc.sock
# Type ctl-b d to detach from the tmux window and leave it running
# in the background.
# Start the tridas shell
> tscsh --socket /var/run/user/10350/tsc.sock
# At the tridas shell prompt enter one of the following:
# --- for 5x5 calorimeter cosmics in Hall-D:
> init /home/streamdaq/tridas/run/datacard_repo/tridas_datacard2022-clondaq3-cosmic-M5EICv1.json
# --- for 5x5 calorimeter beam in Hall-D:
> init /home/streamdaq/tridas/run/datacard_repo/tridas_datacard2022-clondaq3-beam-M5EICv1.json
# --- for 3x3 calorimeter in Hall-B:
> init /home/streamdaq/tridas/run/datacard_repo/tridas_datacard2022-clondaq5-M5EICv1.json
# Then, for all run types, enter:
> configure
> start
Start Run
# Back in the VNC window on the RunControl GUI: - Click 'Prestart', connection should be established from test2vtp-s2 to clondaq5 port 7002 - Click 'Go' - data should be streaming to clondaq5 port 7002
End Run
# To End the run: # In tscsh tridas shell > stop > reset # In VNC RunControl GUI - Click 'Stop' - Click 'Abort' # n.b. remember to click 'Configure' and then 'Download' if you wish to take another run!
Analyzing the Data
Output data files are written to a directory/filename format specified in the datacard.json configuration in the "EM" section by the "PT_NAME_TEMPLATE" field. For the cosmic runs in the Hall-B Counting house, they will have names like:
/data/tridas/EIC_3x3_R00000235_F00000003.pt
You can check the file format is good using PTFileAnalyser like this:
cd /usr/clas12/streaming/work/2022.12.15.JANA2_TriDAS/tridas-core ./build/tools/PTFileAnalyser --ptfile /data/tridas/EIC_3x3_R00000255_F00000000.pt
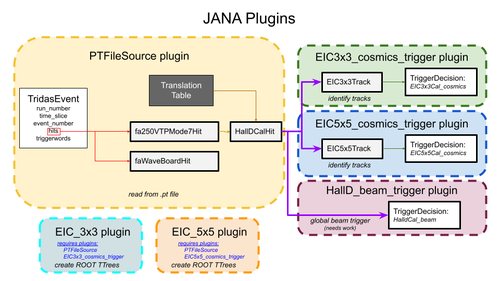
For the 3x3 calorimeter, you can use JANA and the EIC_3x3 plugin to create a ROOT file with some histograms and trees containing the data. Note that this requires the PTFileSource and EIC_3x3_cosmics_trigger plugins also be attached. The former for reading .pt file and translation table and the latter for identifying tracks and triggers in the event. The following will create a file named EIC_3x3.root.
jana -Pplugins=PTFileSource,EIC_3x3_cosmics_trigger,EIC_3x3 /data/tridas/EIC_3x3_R00000321_F00000000.pt
Note: the source for the EIC_3x3 plugin can be found on the web here and on clondaq5 in the directory:
/usr/clas12/streaming/work/2022.12.15.JANA2_TriDAS/streamingReco/src/plugins/EIC_3x3
The other plugins are also in subdirectories of plugins.
Software
Work is being done using the streamdaq account on clondaq5.
NOTE: The .bashrc file for the streamdaq account has been updated to point to the current software versions. The following is currently not needed to complete the environment setup.
source /usr/clas12/streaming/work/2022.12.15.JANA2_TriDAS/setenv.sh
Some software built specifically for this test can be found in the /usr/clas12/streaming/work/2022.12.15.JANA2_TriDAS directory. Not all of it is placed there though. Commands used to build most of the packages are recorded in the /usr/clas12/streaming/work/2022.12.15.JANA2_TriDAS/README file.
Some notes describing how these pieces fit together from 2020 when this was first set up can be seen here:
https://github.com/andrea-celentano/jtridas/blob/master/README.JANA
Compiler
- 9.3.1 (from devtoolset-9)
- C++17 standard
TriDAS
This is the TriDAS source and is checked out on the latest master. It was needed since TriggerInterface.h (and possibly other files) needed by the jtridas plugin were not present in the binary distribution that was installed.
- /usr/clas12/streaming/work/2022.12.15.JANA2_TriDAS/tridas-core
tscsh
The tridas shell tscsh has also been built from the latest master. It is built in the directory below which automatically added to PATH in .bashrc.
- /usr/clas12/streaming/work/2022.12.15.JANA2_TriDAS/tscell/build
jtridas
This is the TriDAS plugin that interfaces to JANA2. TriDAS will load this plugin and, in turn, this will instantiate a JANA2 JApplication that will then load all of the JANA2 plugins. It is responsible for interfacing TriDAS and JANA2.
- /usr/clas12/streaming/work/2022.12.15.JANA2_TriDAS/jtridas
NOTE: This plugin needs to be at least symlinked in the directory where tridas looks for plugins as specified in the datacard.json file. (This is the "PLUGINS_DIR" field in the "TCPU" section.) The following does this for this beam test:
ln -s /usr/clas12/streaming/work/2022.12.15.JANA2_TriDAS/jtridas/build_vscode/libTrigJANA.so /usr/clas12/streaming/work/2022.12.15.JANA2_TriDAS/tridas-core/install/trigger_plugins
streamingReco
This has a number of things specific to this project, but mainly houses the JANA2 plugins. Configuration files for using the cmake build system have been added for this test. The older scons scripts are still there, but moving forward, cmake will be the preferred build system. This is primarily due to cmake being much more prevalent now and interfacing better with modern IDEs.
- /usr/clas12/streaming/work/2022.12.15.JANA2_TriDAS/streamingReco
ROOT
ROOT was built specifically for this test using the above compiler and C++ standard
- /usr/clas12/streaming/root/root-6.26.10
CCDB
CCDB is the JLab Calibrations Constants Database. Version 1.06.09 from github is checked out here.
- /usr/clas12/streaming/ccdb_versions/v1.06.09
JANA2
The latest tagged version of JANA2 v2.0.8 was built for this test. Note that the install directory is given below, but the full source can be found one directory up.
- /usr/clas12/streaming/JANA2-v2.0.8/install
git
The tridas-core CMakeLists.txt runs git internally with the -C option which is only available in newer versions of git. Thus, the latest version (2.39.1 as of now) was built here.
- /usr/clas12/streaming/git/git-2.39.1
cmake
The tridas-core CMakeLists.txt requires version 3.20 or higher of cmake. A precompiled version was downloaded and installed here.
- /usr/clas12/streaming/cmake-3.26.0-rc1-linux-x86_64
libssh2
TriDAS requires Libssh2 version 1.9. A previous build of this from 2020 exists in /usr/clas12/streaming/libssh2-1.9.0 but was apparently built as a static library which caused complaints when linking that we need to rebuild it with -fPIC. This was done and the shared library version placed here. (The older build with the static library was left alone).
- /usr/clas12/streaming/work/2022.12.15.JANA2_TriDAS/libssh2
VSCode
VSCode is an IDE for software development. A version was installed here in case it is useful. The setenv.sh script above will setup an alias to it so you only need to type "code" to start it up. Using VSCode is not required, but may help in some cases.
- /usr/clas12/streaming/work/2022.12.15.JANA2_TriDAS/VSCode-linux-x64
- Important directories
- /usr/clas12/streaming/work/2022.12.15.JANA2_TriDAS
Useful links
- Run Spreadsheet
- New data format (better suited for VTP)
- streamingReco GitHub repository
- jtridas GitHub Repository
- Data located at: /data/tridas/EIC*